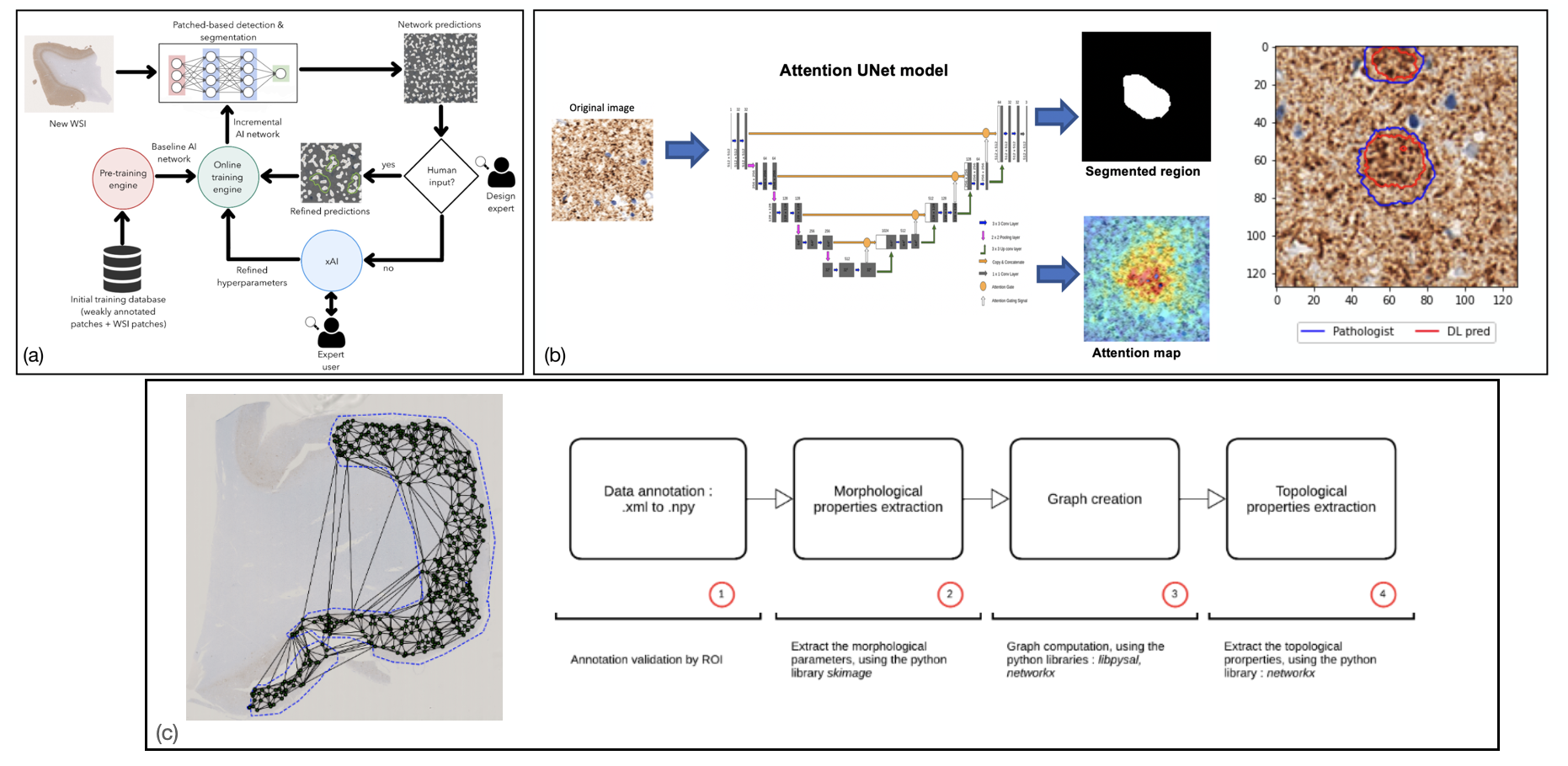
The project aims at developing algorithms for fully automated localization and characterization of tau and Aß aggregates in histological images of the human brain. The topography and morphology of the aggregates are heterogeneous with Aß accumulation taking the form of focal deposits or diffuse plaques and tau lesions forming neurofibrillary tangles.
Within this project, multiple research themes are being explored such as:
- Use of deep learning models with explainability features for segmentation of brain tissue images.
- Development of a cloud based platform for visualization and deep learning based annotation creation for whole slide images
- Use of graph models for representation of morphology and topology of tau objects
GitHub:
https://github.com/stratifIADPapers:
- Gabriel Jimenez, Pablo Mas, Anuradha Kar, Julien Peyrache, Lea Ingrassia, Susana Boluda, Benoît Delatour, Lev Stimmer, Daniel Racoceanu, "A meta-graph approach for analyzing whole slide histopathological images of human brain tissue with Alzheimer's disease biomarkers," Proc. SPIE 12471, Medical Imaging 2023: Digital and Computational Pathology, 124711T , San Diego, USA.
- G. Jimenez et al., “Visual Deep Learning-Based Explanation for Neuritic Plaques Segmentation in Alzheimer’s Disease Using Weakly Annotated Whole Slide Histopathological Images,” In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. Lecture Notes in Computer Science, vol 13432. Springer, Cham, 2022, pp. 336–344.
- Kar, A, Improving segmentation quality with fusion based post-processing: An assessment with 3D deep learning and classical segmentation pipelines, CVPR-CVMI 2022 (highlights) at the 7th IEEE Workshop on Computer Vision for Microscopy Image Analysis (CVMI) at the 2022 IEEE CVPR conference, June 19, 2022.
- Garay, G.J., Kar, A., Ounissi, M., Stimmer, L., Delatour, B. and Racoceanu, D. (2022), Interpretable Deep Learning in Computational Histopathology for refined identification of Alzheimer’s Disease biomarkers. Alz. Dement., 18: e065363. - AAIC 2022, San Diego, USA.
- Kar A., Jimenez G., Ounissi M., Stimmer L., Delatour B., Racoceanu D., A deep learning framework for stratification of Alzheimer’s disease patients using whole slide histopathological brain tissue images, European Conference on Digital Pathology - ECDP 2022, Berlin, Germany.
- G. Jiménez,A. Kar,M. Ounissi,D. Racoceanu, "Empowering Researchers for Understanding Alzheimer’s Disease using Explainable AI", AI4 Health Winter School (virtual) 2022, 10-14th January 2022.





